00040-Drug Repurposing for Covid-19 via Disease-ubuntu
前言
Drug Repurposing Knowledge Graph (DRKG) is a comprehensive biological knowledge graph relating genes, compounds, diseases, biological processes, side effects and symptoms. DRKG includes information from six existing databases including DrugBank, Hetionet, GNBR, String, IntAct and DGIdb, and data collected from recent publications particularly related to Covid19. It includes 97,238 entities belonging to 13 entity-types; and 5,874,261 triplets belonging to 107 edge-types. These 107 edge-types show a type of interaction between one of the 13 entity-type pairs (multiple types of interactions are possible between the same entity-pair), as depicted in the figure below. It also includes a bunch of notebooks about how to explore and analysis the DRKG using statistical methodologies or using machine learning methodologies such as knowledge graph embedding.
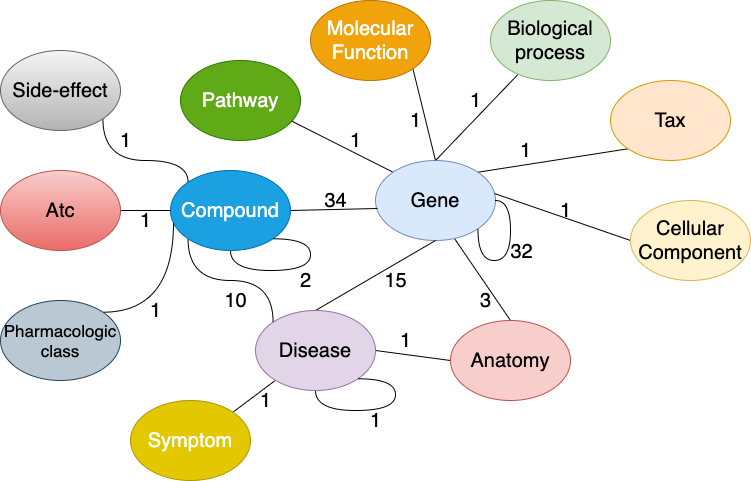
Figure: Interactions in the DRKG. The number next to an edge indicates the number of relation-types for that entity-pair in DRKG.
原项目地址: https://github.com/gnn4dr/DRKG .
操作系统:Ubuntu 20.04.5 LTS
参考文档
配置环境
下载 DRKG
为了分析 DRKG, 可以直接通过下面的命令下载 DRKG.
1 | wget https://dgl-data.s3-us-west-2.amazonaws.com/dataset/DRKG/drkg.tar.gz |
(不推荐) 如果直接使用项目仓库中的 notebooks , 不需要手动的下载 DRKG, 因为会自动下载 DRKG.
当你解压 drkg.tar.gz, 将会发现下面的文件:
1 | ./drkg.tsv |
版本
Python 版本 (参考):
1 | python --version |
Python 包版本 (参考):
1 | Package Version |
真正的配置流程
最简单的方法就是直接 git clone 项目仓库.
1 | git clone https://github.com/gnn4dr/DRKG.git |
直接在浏览器的地址栏输入 https://dgl-data.s3-us-west-2.amazonaws.com/dataset/DRKG/drkg.tar.gz, 下载到上面的 data 目录中, 然后解压, 结果如下.
1 | tree data/ |
说明
本文介绍的内容是项目的 DRKG/drug_repurpose/COVID-19_drug_repurposing.ipynb 的内容, 只是额外的打印了一些帮助输出.
COVID-19_drug_repurposing.ipynb shows how to do drug repurposing for Covid-19 by predicting links between the disease entities and the drug entitites in the DRKG. The target disease entities are listed in the notebook and the candidate drug entities are listed in infer_drug.tsv. The drugs are all from Drugbank and we exclude drugs with molecule weight less than 250 daltons which results in 8104 candidates. Two edge types are chosen here: Hetionet::CtD::Compound:Disease and GNBR::T::Compound:Disease, which represent the treatment relationship between a certain drug for a disease. To evaluate the repurposed drugs, we compare them with the clinical drugs as there is no treatment for Covid-19 right now. The list of clinical drugs are shown in COVID19_clinical_trial_drugs.tsv which is collected from http://www.covid19-trials.com/.
COVID-19 Drug Repurposing via disease-compounds relations
This example shows how to do drug repurposing using DRKG even with the pretrained model.
Collecting COVID-19 related disease
At the very beginning we need to collect a list of disease of Corona-Virus(COV) in DRKG. We can easily use the Disease ID that DRKG uses for encoding the disease. Here we take all of the COV disease as target.
1 | COV_disease_list = [ |
1 | len(COV_disease_list) |
34
1 | COV_disease_list[:3] |
['Disease::SARS-CoV2 E', 'Disease::SARS-CoV2 M', 'Disease::SARS-CoV2 N']
Candidate drugs
Now we use FDA-approved drugs in Drugbank as candidate drugs. (we exclude drugs with molecule weight < 250) The drug list is in infer_drug.tsv.
1 | import csv |
1 | len(drug_list) |
8104
1 | drug_list[:3] |
['Compound::DB00605', 'Compound::DB00983', 'Compound::DB01240']
Treatment relation
Two treatment relations in this context
1 | treatment = ['Hetionet::CtD::Compound:Disease','GNBR::T::Compound:Disease'] |
1 | treatment |
['Hetionet::CtD::Compound:Disease', 'GNBR::T::Compound:Disease']
Get pretrained model
We can directly use the pretrianed model to do drug repurposing.
1 | import numpy as np |
1 | entity_idmap_file = '../data/drkg/embed/entities.tsv' |
Get embeddings for diseases and drugs
1 | # Get drugname/disease name to entity ID mappings |
1 | len(disease_ids),len(drug_ids),len(treatment_rid) |
(34, 8104, 2)
1 | disease_ids[:3],drug_ids[:3],treatment_rid |
([9079, 9085, 9110], [9475, 11010, 7486], [68, 35])
1 | # Load embeddings |
1 | disease_ids[:3],drug_ids[:3],treatment_rid |
(tensor([9079, 9085, 9110]), tensor([ 9475, 11010, 7486]), tensor([68, 35]))
1 | drug_emb.shape |
torch.Size([8104, 400])
Drug Repurposing Based on Edge Score
We use following algorithm to calculate the edge score. Note, here we use logsigmiod to make all scores < 0. The larger the score is, the stronger the will have with .
When doing drug repurposing, we only use the treatment related relations.
1 | import torch.nn.functional as fn |
1 | scores.shape, dids.shape, 2*34*8104 |
(torch.Size([551072]), torch.Size([551072]), 551072)
1 | # sort scores in decending order |
((551072,), (551072,), 551072)
Now we output proposed treatments
1 | _, unique_indices = np.unique(dids, return_index=True) |
We select top K relevent drugs according the edge score.
1 | for i in range(topk): |
Compound::DB00811 -0.21416780352592468
Compound::DB00993 -0.8350887298583984
Compound::DB00635 -0.8974790573120117
Compound::DB01082 -0.985488772392273
Compound::DB01234 -0.9984012842178345
Compound::DB00982 -1.0160716772079468
Compound::DB00563 -1.0189464092254639
Compound::DB00290 -1.0641062259674072
Compound::DB01394 -1.080676555633545
Compound::DB01222 -1.084547519683838
Compound::DB00415 -1.0853973627090454
Compound::DB01004 -1.096669316291809
Compound::DB00860 -1.1004788875579834
Compound::DB00681 -1.1011555194854736
Compound::DB00688 -1.1256868839263916
Compound::DB00624 -1.1428292989730835
Compound::DB00959 -1.1618409156799316
Compound::DB00115 -1.186812400817871
Compound::DB00091 -1.1906721591949463
Compound::DB01024 -1.2051165103912354
Compound::DB00741 -1.2147064208984375
Compound::DB00441 -1.2320411205291748
Compound::DB00158 -1.2346546649932861
Compound::DB00499 -1.252516746520996
Compound::DB00929 -1.2730495929718018
Compound::DB00770 -1.282552719116211
Compound::DB01331 -1.2960493564605713
Compound::DB00958 -1.296778917312622
Compound::DB02527 -1.3034359216690063
Compound::DB00196 -1.3053343296051025
Compound::DB00537 -1.3131842613220215
Compound::DB00644 -1.3131849765777588
Compound::DB01048 -1.3267205953598022
Compound::DB00552 -1.3272082805633545
Compound::DB00328 -1.3286100625991821
Compound::DB00171 -1.3300385475158691
Compound::DB01212 -1.33307683467865
Compound::DB09093 -1.3382985591888428
Compound::DB00783 -1.3385637998580933
Compound::DB09341 -1.3396947383880615
Compound::DB00558 -1.3425898551940918
Compound::DB05382 -1.3575100898742676
Compound::DB01112 -1.3584487438201904
Compound::DB00515 -1.3608112335205078
Compound::DB01101 -1.3815491199493408
Compound::DB01165 -1.3838152885437012
Compound::DB01183 -1.3862131834030151
Compound::DB00815 -1.3863469362258911
Compound::DB00755 -1.3881793022155762
Compound::DB00198 -1.3885042667388916
Compound::DB00480 -1.3935296535491943
Compound::DB00806 -1.3996552228927612
Compound::DB01656 -1.3999735116958618
Compound::DB00759 -1.4046530723571777
Compound::DB00917 -1.4116041660308838
Compound::DB01181 -1.4148895740509033
Compound::DB01039 -1.4176596403121948
Compound::DB00512 -1.4207416772842407
Compound::DB01233 -1.4211865663528442
Compound::DB11996 -1.4257901906967163
Compound::DB00738 -1.4274098873138428
Compound::DB00716 -1.4327492713928223
Compound::DB03461 -1.437927484512329
Compound::DB00591 -1.4404346942901611
Compound::DB01327 -1.4408750534057617
Compound::DB00131 -1.4446901082992554
Compound::DB00693 -1.4460757970809937
Compound::DB00369 -1.4505729675292969
Compound::DB04630 -1.453115463256836
Compound::DB00878 -1.4564695358276367
Compound::DB08818 -1.4633687734603882
Compound::DB00682 -1.4691758155822754
Compound::DB01068 -1.470010757446289
Compound::DB00446 -1.4720206260681152
Compound::DB01115 -1.4729849100112915
Compound::DB00355 -1.4770021438598633
Compound::DB01030 -1.4850695133209229
Compound::DB00620 -1.497349500656128
Compound::DB00396 -1.497694492340088
Compound::DB01073 -1.498704433441162
Compound::DB00640 -1.502620816230774
Compound::DB00999 -1.503427505493164
Compound::DB01060 -1.5043613910675049
Compound::DB00493 -1.5072377920150757
Compound::DB01240 -1.5090980529785156
Compound::DB00364 -1.5099471807479858
Compound::DB01263 -1.5119924545288086
Compound::DB00746 -1.5130668878555298
Compound::DB00718 -1.5183119773864746
Compound::DB01065 -1.5207159519195557
Compound::DB01205 -1.521277904510498
Compound::DB01137 -1.5229606628417969
Compound::DB08894 -1.5239675045013428
Compound::DB00813 -1.5308716297149658
Compound::DB01157 -1.5316542387008667
Compound::DB04570 -1.5430858135223389
Compound::DB00459 -1.550320029258728
Compound::DB01752 -1.554166555404663
Compound::DB00775 -1.555970549583435
Compound::DB01610 -1.5563467741012573
Check Clinial Trial Drugs
There are seven clinial trial drugs hit in top100. (Note: Ribavirin exists in DRKG as a treatment for SARS)
1 | clinical_drugs_file = './COVID19_clinical_trial_drugs.tsv' |
[0] Ribavirin -0.21416780352592468
[4] Dexamethasone -0.9984012842178345
[8] Colchicine -1.080676555633545
[16] Methylprednisolone -1.1618409156799316
[49] Oseltamivir -1.3885042667388916
[87] Deferoxamine -1.5130668878555298
32
附录
上面脚本中下载 DRKG 的函数是在 DRKG/utils/utils.py 中.
函数定义如下:
1 | import os |
结语
第四十篇博文写完,开心!!!!
今天,也是充满希望的一天。