00041-DRKG 原论文学习笔记-ubuntu
前言
DRKG 是一个由 Amazon Web Services Shanghai AI Lab 联合多家科研机构开源的综合生物知识图谱, 涉及基因、药物、疾病、生物学过程、副作用和症状. DRKG 包括 DrugBank、Hetionet、GNBR、String、IntAct 和 DGIdb 六个现有数据库的信息, 以及从近期新冠病毒的相关医学文献中挖掘的数据. DRKG 知识图谱包含属于 13 种实体类型的 97,238 个实体, 以及属于 107 种关系类型的 5,874,261 个三元组. 同一实体对之间可能存在多种交互类型.
DRKG 原论文链接: DRKG Drug Repurposing Knowledge Graph.
原仓库地址: https://github.com/gnn4dr/DRKG .
操作系统:Ubuntu 20.04.5 LTS
参考文档
- B. Kotnis and V. Nastase, “Analysis of the impact of negative sampling on link prediction in knowledge graphs,” 2017.
项目仓库 - Drug Repurposing Knowledge Graph (DRKG)
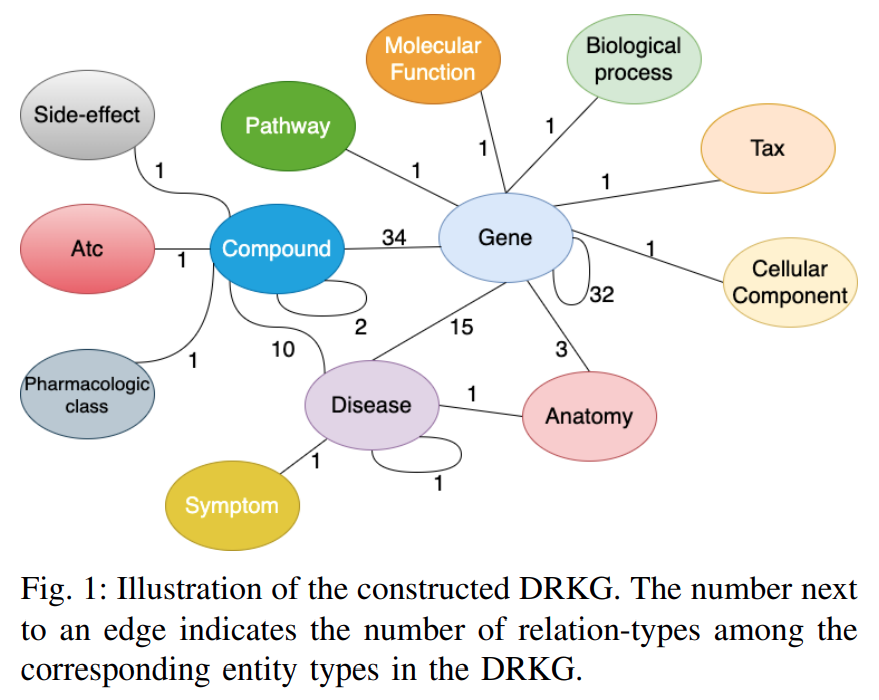
药物再利用知识图谱 (Drug Repurposing Knowledge Graph, DRKG)1 是一个涉及基因、药物、疾病、生物过程、副作用和症状的综合生物知识图谱. DRKG 包括来自 DrugBank、Hetionet、GNBR、String、IntAct 和 DGIdb 等六个现有数据库的信息, 以及从最近发表的 Covid19 出版物中收集的数据. 它包含属于 13 种实体类型的 97,238 个实体; 以及属于 107 种关系类型的 5,874,261 个三元组. 这 107 种关系类型显示了 13 种实体类型之间的交互类型(同一种实体对之间可能存在多种交互类型), 如下图所示. 它还包括很多关于如何使用统计方法或使用机器学习方法(如知识图谱嵌入)探索和分析 DRKG 的 notebooks.
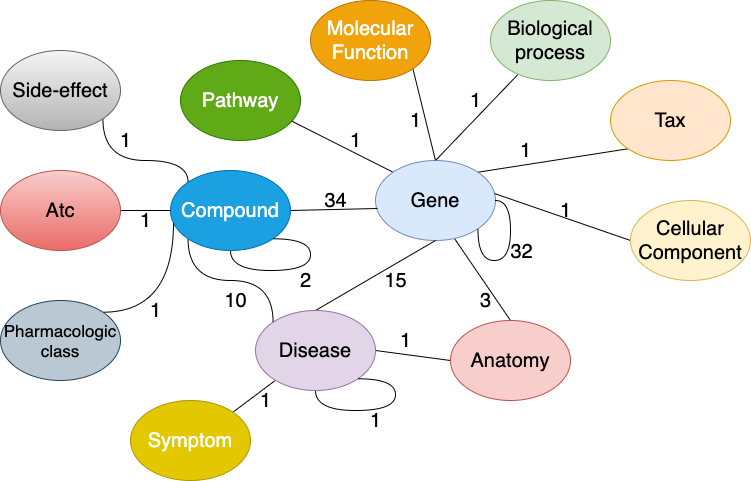
Figure: Interactions in the DRKG. The number next to an edge indicates the number of relation-types for that entity-pair in DRKG.
Statistics of DRKG
The type-wise distribution of the entities in DRKG and their original data-source(s) is shown in following table.
| Entity type | Drugbank | GNBR | Hetionet | STRING | IntAct | DGIdb | Bibliography | Total Entities |
|---|---|---|---|---|---|---|---|---|
| Anatomy | - | - | 400 | - | - | - | - | 400 |
| Atc | 4,048 | - | - | - | - | - | - | 4,048 |
| Biological Process | - | - | 11,381 | - | - | - | - | 11,381 |
| Cellular Component | - | - | 1,391 | - | - | - | - | 1,391 |
| Compound | 9,708 | 11,961 | 1,538 | - | 153 | 6,348 | 6,250 | 24,313 |
| Disease | 1,182 | 4,746 | 257 | - | - | - | 33 | 5,103 |
| Gene | 4,973 | 27,111 | 19,145 | 18,316 | 16,321 | 2,551 | 3,181 | 39,220 |
| Molecular Function | - | - | 2,884 | - | - | - | - | 2,884 |
| Pathway | - | - | 1,822 | - | - | - | - | 1,822 |
| Pharmacologic Class | - | - | 345 | - | - | - | - | 345 |
| Side Effect | - | - | 5,701 | - | - | - | - | 5,701 |
| Symptom | - | - | 415 | - | - | - | - | 415 |
| Tax | - | 215 | - | - | - | - | - | 215 |
| Total | 19,911 | 44,033 | 45,279 | 18,316 | 16,474 | 8,899 | 9,464 | 97,238 |
The following table shows the number of triplets between different entity-type pairs in DRKG for DRKG and various datasources.
| Entity-type pair | Drugbank | GNBR | Hetionet | STRING | IntAct | DGIdb | Bibliography | Total interactions |
|---|---|---|---|---|---|---|---|---|
| (Gene, Gene) | - | 66,722 | 474,526 | 1,496,708 | 254,346 | - | 58,629 | 2,350,931 |
| (Compound, Gene) | 24,801 | 80,803 | 51,429 | - | 1,805 | 26,290 | 25,666 | 210,794 |
| (Disease, Gene) | - | 95,399 | 27,977 | - | - | - | 461 | 123,837 |
| (Atc, Compound) | 15,750 | - | - | - | - | - | - | 15,750 |
| (Compound, Compound) | 1,379,271 | - | 6,486 | - | - | - | - | 1,385,757 |
| (Compound, Disease) | 4,968 | 77,782 | 1,145 | - | - | - | - | 83,895 |
| (Gene, Tax) | - | 14,663 | - | - | - | - | - | 14,663 |
| (Biological Process, Gene) | - | - | 559,504 | - | - | - | - | 559,504 |
| (Disease, Symptom) | - | - | 3,357 | - | - | - | - | 3,357 |
| (Anatomy, Disease) | - | - | 3,602 | - | - | - | - | 3,602 |
| (Disease, Disease) | - | - | 543 | - | - | - | - | 543 |
| (Anatomy, Gene) | - | - | 726,495 | - | - | - | - | 726,495 |
| (Gene, Molecular Function) | - | - | 97,222 | - | - | - | - | 97,222 |
| (Compound, Pharmacologic Class) | - | - | 1,029 | - | - | - | - | 1,029 |
| (Cellular Component, Gene) | - | - | 73,566 | - | - | - | - | 73,566 |
| (Gene, Pathway) | - | - | 84,372 | - | - | - | - | 84,372 |
| (Compound, Side Effect) | - | - | 138,944 | - | - | - | - | 138,944 |
| Total | 1,424,790 | 335,369 | 2,250,197 | 1,496,708 | 256,151 | 26,290 | 84,756 | 5,874,261 |
Download DRKG
为了分析 DRKG, 你可以直接使用下面的命令下载 DRKG:
1 | wget https://dgl-data.s3-us-west-2.amazonaws.com/dataset/DRKG/drkg.tar.gz |
(不推荐) 如果你使用原仓库提供的 notebooks, 不需要手动下载 DRKG.
解压 drkg.tar.gz, 会得到下面的文件:
1 | ./drkg.tsv |
DRKG dataset
数据集包含四部分:
-
drkg.tsv, 一个包含原始 DRKG 三元组 (格式为 (h, r, t)) 的 tsv 文件.
-
embed, 一个包含使用全部 drkg.tsv 文件为训练集的预训练知识图谱嵌入和 molecule SMILES 的预训练 GNN 分子嵌入的目录.
-
entity2src.tsv, DRKG 中的实体到源数据的映射文件.
-
relation_glossary.tsv, DRKG 关系类型的解释文件, 以及相关的源信息.
Pretrained DRKG embedding
这些 DRKG 嵌入是使用 400 维度的 TransE_l2 模型训练的, 有 4 个文件:
-
DRKG_TransE_l2_entity.npy, 二进制的 NumPy 的数据, 存储实体嵌入.
-
DRKG_TransE_l2_relation.npy, 二进制 NumPy 的数据, 存储关系嵌入.
-
entities.tsv, 映射实体名到实体 ID.
-
relations.tsv, 映射关系名到关系 ID.
可以使用 np.load 函数单独加载实体嵌入和关系嵌入:
1 | import numpy as np |
Pretrained Molecule Embedding
DRKG 也提供了 DrugBank 的大多数小分子药物的分子嵌入 (使用预训练的 GNN). Strategies for Pre-training Graph Neural Networks 将监督的分子性质预测与自我监督学习结合, 开发了多种方法用于预训练基于 GNN 的分子表示. 下面的 notebooks 使用 DGL-LifeSci 计算了四种分子嵌入的变体:
-
mol_contextpred.npy: 预测分子子图的周围图结构的预训练模型.
-
mol_infomax.npy:最大化局部节点表示和全局图谱表示间的交互信息的预训练模型.
-
mol_edgepred.npy: 鼓励邻近节点有类似的表示和强迫不相干的节点有不同的表示的预训练模型.
-
mol_masking.npy: 预测随机遮蔽节点和边的属性的预训练模型.
Tools to analyze DRKG
分析 DRKG 的深度学习框架, 包括 DGL (一个图神经网络的框架) 和 DGL-KE (一个计算知识图谱嵌入的库).
Install PyTorch
所有的 notebooks 都使用 PyTorch 作为深度学习后端. 可以去 Pytorch 官网 安装其他版本.
1 | sudo pip3 install torch==1.5.0+cu101 torchvision==0.6.0+cu101 -f https://download.pytorch.org/whl/torch_stable.html |
Install DGL
使用下面命令安装 DGL (一个图神经网络框架), 它会安装支持 CUDA 的 DGL.
1 | sudo pip3 install dgl-cu101 |
可以前往 Install DGL 安装 DGL 的其他版本.
Install DGL-KE
如果你想用这些位于 embedding_analysis 的 notebooks (Train_embeddings.ipynb or Edge_score_analysis.ipynb) 训练模型, 你需要安装 DGL and DGL-KE, DGL-KE 能够和 DGL (版本 >= 0.4.3, either CPU or GPU) 一起工作.
1 | sudo pip3 install dglke |
Notebooks for analyzing DRKG
Basic Graph Analysis of DRKG
为了评估一对关系类型之间的结构相似性, 通过重叠系数计算两个关系类型的 Jaccard 相似系数和重叠.
Knowledge Graph Embedding Based Analysis of DRKG
通过学习一个利用 距离的 TransE KGE 模型分析 DRKG. 由于 DRKG 结合了来自不同数据源的信息, 我们希望验证使用知识图谱嵌入技术可以生成有意义的实体和关系嵌入.
将数据集分成训练集 (90%), 验证集 (5%), 测试集 (5%), 使用下面的 notebook 训练 KGE 模型:
最终, 得到了 DRKG 实体和关系的嵌入. 可以使用下面的 notebooks 进行嵌入分析.
-
Relation_similarity_analysis.ipynb, 分析生成的关系嵌入相似性.
-
Entity_similarity_analysis.ipynb, 分析生成的实体嵌入相似性.
-
Edge_score_analysis.ipynb, 评估学习到的 KGE 模型能否预测 DRKG 的关系 (edges).
-
Edge_similarity_based_on_link_recommendation_results.ipynb, 评估不同关系类型之间的预测链接的相似程度.
Drug Repurposing Using Pretrained Model for COVID-19
在使用预训练模型进行 COVID-19 的药物再利用的例子中, 直接使用 DRKG 的预训练模型推荐了治疗 COVID-19 的 100 个药物. 对应的 notebook:
DRKG with DGL
下面的 notebook 提供了利用 DGL 从 DRKG 中构建异构图的示例, 以及一些异构图的查询示例:
Additional Information for DrugBank
drugbank_info 包括来自 DrugBank 药物的额外信息, 包含药物的类型, 重量和小分子药物的 SMILES.
Licence
This project is licensed under the Apache-2.0 License. However, the DRKG integrates data from many resources and users should consider the licensing of each source (see this table) . Authors apply a license attribute on a per node and per edge basis for sources with defined licenses.
Cite
Please cite the dataset if you use this code and data in your work.
1 | @misc{drkg2020, |
DRKG 原论文学习笔记
论文题目: DRKG - Drug Repurposing Knowledge Graph for Covid-19
Covid-19 大流行的时间表表明迫切需要快速开发针对新疾病的有效治疗方法. 药物再利用 (Drug Repurposing, DR) 是一种来自现有药物的药物发现策略, 与从头发现药物相比, 该策略显著缩短了时间并降低了成本. 本文详细介绍了创建一个与基因、药物、疾病、生物过程、副作用和症状相关的综合生物知识图谱的工作, 该知识图谱被称为药物再利用知识图谱 (Drug Repurposing Knowledge Graph, DRKG). DRKG 包括来自不同数据库的信息, 例如 DrugBank, String, GNBR, 以及从最近出版物中收集的数据, 特别是与 Covid19 相关的数据. DR 可以通过预测基因和化合物之间的新相互作用来解决, 这可以表述为 DRKG 上的链接预测任务. 提供了分析构建的 DRKG, 过滤噪声链路和节点的方法. 图机器学习模型可以利用 DRKG 进行 DR, 并预测现有药物是否成功抑制与 Covid-19 宿主蛋白相关的某些途径. 使用最先进的知识图嵌入模型, 学习 DRKG 中实体和关系的嵌入. 还进行分析以验证图结构和学习嵌入是否具有高质量. 最后, 利用在 DRKG 上学习到的 KGE, 评估了新冠肺炎的药物再利用. 结果证实, 临床试验中使用的几种药物被确定为可能的候选药物. 最后, 通过将 DRKG 的药物再利用结果与组成数据库的结果进行比较, 确认值得构建一个全面的 DRKG.
The methods presented in this paper are implemented in
the efficient deep graph learning(DGL) library ( https://www.dgl.ai/ ).The
DRKG,entity & relation embeddings, andthe source codefor the analysis presented in this paper ispublicly available( https://github.com/gnn4dr/DRKG/ ).
介绍
2019 年冠状病毒病 (COVID-19) 国际突发公共卫生事件说明了提高发现新疗法的效率和速度的紧迫性. 不幸的是, 从头药物发现是一个漫长而昂贵的过程, 通常需要 10-15 年, 每种 FDA 批准的新药的成本超过 26 亿美元.
An alternate approach that can dramatically reduce the time to discover new treatments is
drug repurposing (DR)(orrepositioning),which seeks to redevelop existing drugs for use in different diseases.
DRleverages the fact thatcommon molecular pathways contribute to different diseasesand hencesome drugs may be reused.It capitalizes on
the existence of detailed information on approved drugs and many abandoned compoundsrelated to theirpharmacology(药理学),formulation(制剂),dose(剂量), andpotential toxicity(潜在毒性).
药物再利用依赖于识别基因和化合物等生物实体之间的新相互作用.
Traditional approachesfor doing that rely oncostly and time-consumingexperimental methodologies.As a result,
several approacheshave been developed that aim toleverage the diverse types of informationthat already exists about thedrugs, theirtargets, and thediseasesin order toreduce the cost and speedup drug repurposing.Among them,
approachesthatrepresent the existing informationin a form ofa knowledge graphand deploy graph-based machine learning techniques based ongraph neural networksandknowledge graph embedding modelshave gained popularity.
贡献
在这个项目中, 构建了一个称为 DRKG 的多样化药物再利用知识图谱, 并提供了一套机器学习工具, 用于加快药物再利用.
We
collect interactionsfrom the followingpublicly-availabledata sources: (i)Drugbank, (ii)Global Network of Biomedical Relationships (GNBR), (iii)Hetionet, (iv)STRING, (v)IntAct, (vi)DGIdb, and (vii)relations from bibliographic sources.
We map
the biological entities of the different databasestoa common ID space, whichallows us to link entities across databases, and wefilter the initial data to remove noisy links and entities.
In total,
DRKGcontains97,055vertices belonging to13types of entities, and5,869,294edges belonging to107types of relations.
In addition,
DRKGcontainsa number of Covid-19 related proteins and genes, asextracted from recent publications.
To analyze DRKG, we
formulateandsolvethe link prediction task using models that computeknowledge graph embedding (KGE).
We
perform analysisto validate thatthe graph structureandthe learned embeddings are of high quality.
Our analysis shows that
similar biological entities and relations have similar embeddingsthat corroborates insights from biology and henceDRKG can be used for developing machine learning models.
Finally, we
used these embeddings to evaluate how well DRKG can be used to identify drugsthat can be repurposed for Covid-19.Our results show that among the highest scoring drugs, there are several drugs undergoing clinical trials.
These results illustrate that
using the DRKG, one can apply machine learning models to predict new linksandfacilitate drug repurposing for novel diseases.
Finally, by comparing
the drug repurposing results of DRKGwiththose of the constituent databases, we confirm hemerits to constructing a comprehensive DRKG.
The
DRKG,entity & relation embeddings, andthe source codefor the analysis presented in this paper is publicly made available ( https://github.com/gnn4dr/DRKG/ ).
背景
定义和符号
A graph is composed of
two sets—the set of nodes (also calledvertices) and the set of edges (also calledarcs).Each edgeconnectsa pair of nodesindicating thatthere is a relation between them.
This relationcan either beundirected, e.g.,capturing symmetric relations between nodes, ordirected,capturing asymmetric relations. Depending onthe edges’ directionality, a graph can bedirectedorundirected.Graphs can be either
homogeneousorheterogeneous. Ina homogeneous graph,all the nodes represent instances of the same typeandall the edges represent relations of the same type. In contrast,in a heterogeneous graph,the nodes and edges can be of different types.Finally, a
graphcan either bea simple graphora multigraph. Ina simple graphthere isonly a single directed edgeconnecting a pair of nodes andit does not have loops. In contrast,in a multigraphthere can bemultiple (directed) edgesbetweenthe same pair of nodesand can alsocontain loops.
These multiple edgesaretypically of different typesand assuch most multigraphs are heterogeneous.
A knowledge graph (KG)is adirected heterogeneous multi-graphwhose node and relation types havedomain-specific semantics.
KGsallow us toencode the knowledge into a form that is human interpretableand amenable to automatedanalysisandinference.
A nodein a knowledge graph representsan entityandan edgerepresentsa relation between two entities.The edges are usually in the form of triplets , each of which indicates that a pair of entities (head) and (tail) are coupled via a relation .
Knowledge graph embeddingsarelow-dimensional representation of entities and relations.
These embeddingscarrythe information of the entities and relationsin the knowledge graph and are widely used in tasks, such asknowledge graph completionandrecommendation.Throughput the paper, we denote
the embedding vector of head entity, tail entity and relationwith , and , respectively;all embeddings have the same dimension sizeof .
知识图谱嵌入 (KGE) 模型
The knowledge graph embeddingsare computed so thatthey satisfy certain properties; i.e., they followa given KGE model. A KGE modeldefines a score functionthatmeasures the distance of two entities relative to its relation typein the low-dimensional embedding space.
This score functionisdefined on the tripletsandduring training it optimizes a loss functionthatmaximizes the scores on triplets that exist in the knowledge graph (positive triplets)andminimizes the scores on triplets that do not exist (negative triplets).
use
the logistic lossforKGE model traininggiven by
where and are
the positive and negative sets of triplets, and if the triplet corresponds to a positive example and otherwise.
The negative tripletsare usuallyconstructedby replacingthe entities or relations in positive tripletswithentities and relations randomly sampled from the knowledge graph.For
a review of negative sampling strategiessee 2.
Many score functionshave been developed totrain knowledge graph embeddingsandTable Iliststhe score functions of some of them.
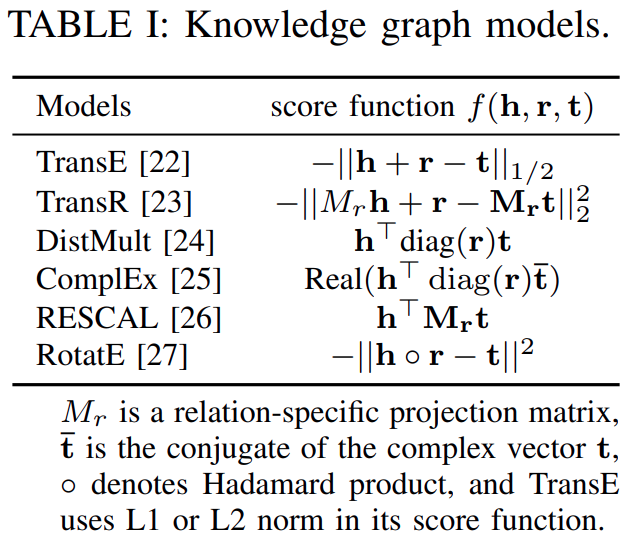
TransEandTransRarerepresentative translational modelsthatexplore the Euclidean distance-based scoring functions, whileDistMult,ComplEx, andRESCALaresemantic matching modelsthatexploit similarity-based scoring functions.
Obtaining embeddings of entities and relationsin the DRKGis beneficial fora number ofdownstream tasks.First,
the embeddings may be used to clean the DRKG from noisy tripletsthat may appear due to erroneous entries in thedata sources.Second,
the embeddings can be used to establish the similarity of different entities and relation typessuch as diseases, drugs, interaction types, and obtain clusters of similar entities.Third,
the embeddings can be used to predict putative new links between the DRKG’s entities, which can contribute toour overall knowledge about the diseasome.For example,
embeddingscan be used todiscover new side-effects of drugsandidentify alternate therapuetic uses of existing drugs(see bellow).
药物再利用知识图谱
药物来源
1)
Drugbank:The DrugBank databaseisa bioinformatics and cheminformatics knowledge basewithrich drug data informationas well ascomprehensive drug target information.used
the latest version (version 5.1.5)which contains13563drug entries.used
Bio2RDF( https://github.com/bio2rdf/bio2rdf-scripts ) toextract the triplets corresponding to the relationsshown inTable IIfrom Drugbank’s original XML format.
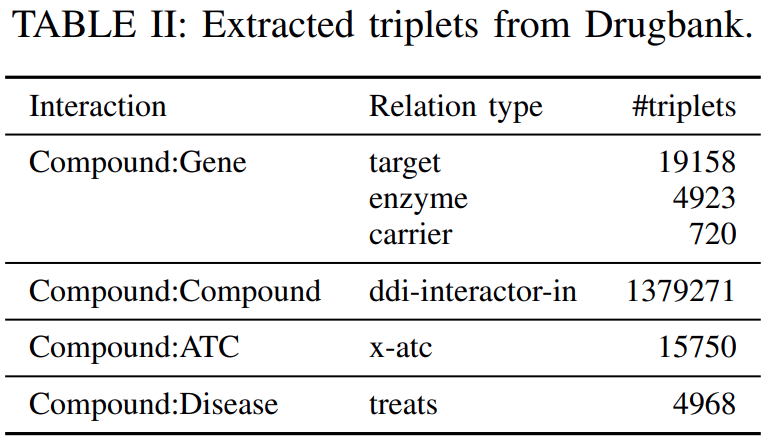
Note that
the treats relation corresponds to Drugbank’s associated conditions.
since the ATC classification is hierarchical, for each tripletinvolving a specific ATC code, we alsoincluded triplets corresponding to the higher-level ATC codes. For example, if(DB09344, x-atc, Atc::C05BB03)is an original triplet, we added the following triplets:(DB09344, x-atc, Atc::C05BB),(DB09344, x-atc, Atc::C05B),(DB09344, x-atc, Atc::C05), and(DB09344, x-atc, Atc::C).
excluded entitiesthat participated inless than 2 tripletsandtripletsconnecting to these entities.In total, we extracted from Drugbank
1419822 tripletsand18729 entitieswhose statistics are shown inTable X.
2)
GNBR:Global Network of Biomedical Relationships (GNBR)usesNCBI’s PubTatorannotations to identify instances ofchemical,gene, anddiseasenames inMedline abstracts, andapplies dependency parsing to find dependency paths between pairs of entities in individual sentences.These dependency paths are grouped into
semantically-related categories, toprovide relations (with confidence) between among enitities that appear together in a sentence.
GNBRincludesgene-gene,gene-disease,drug-gene, anddrug-diseaseinteractions.
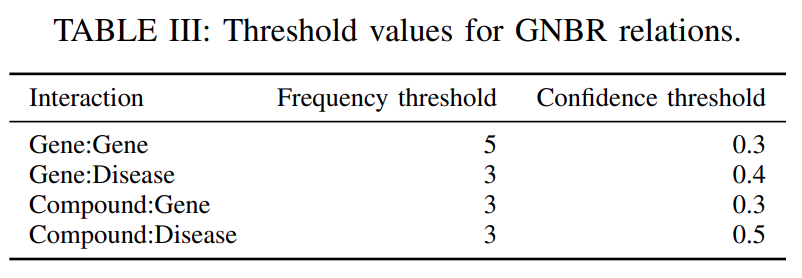
To eliminate the entity pairs co-occurring by chance, we only considered
the pairs that co-occur in the same sentence more than a threshold number of times(column’frequency threshold’in Table III).Next, to find all relations between the pair of entities, we
aggregated the confidence for the pair related by a particular relation, over all the sentences in which that pair of entities co-occurs.We again
selected only those relations whose confidence is more than a threshold(column’confidence threshold’inTable III), so as toremove noisy relationsoccurring by chance.Further, we also
remove all the relations from a gene to itself.
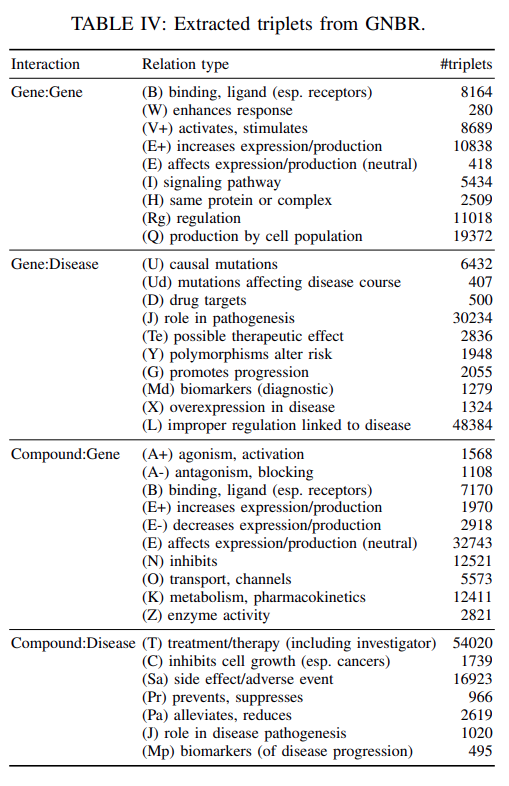
Our final processed dataset extracted from GNBR contains
66722 gene-gene interactions,95400 gene-disease interactions,80803 drug-gene interactions, and77782 drug-disease interactions.
The statistics of the extracted relationsare shown inTable IV
3)
Hetionet:Hetionet is a heterogeneous information network of biomedical knowledge assembled from 29 different databasesrelatinggenes,compounds,diseasesand other.
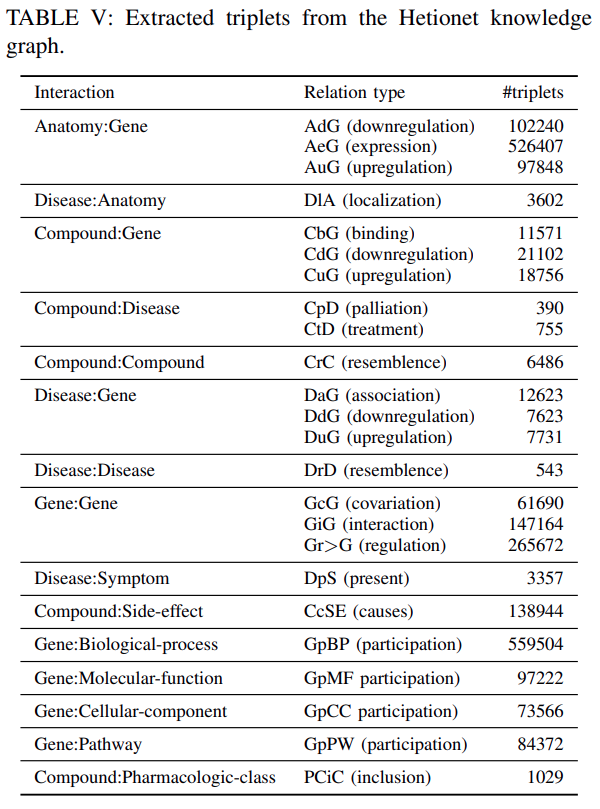
We extracted
2250197 triplets from 24 relation typesas shown inTable Vand45279 entities belong to 11 entities typesas shown inTable X.
4)
STRING:STRING is a database of established and predicted protein-protein interactions.The interactions include
direct (physical)andindirect (functional)associations and are extracted fromcomputational prediction,knowledge transferbetween organisms, and interactions aggregated from other databases.
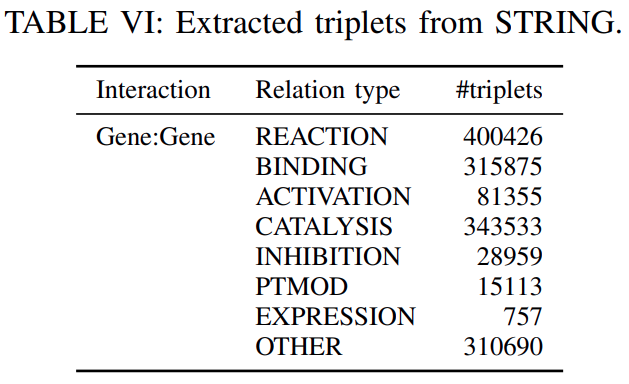
We extracted the interactions
whose score is greater than or equal to 0.6, resulting in1496708 triplets from 7 relation typesas shown inTable VIand18316 gene entities.
5)
IntAct:IntActis an open source database that containsmolecular interaction data.IntAct provides
gene to geneas well asgene to chemical compoundsinteractions.

The extracted relations and entitiesare inTables VIIand X.
6)
DGIdb:DGIdbisa drug–gene interaction databasethatconsolidates,organizesandpresentsdrug–gene interactions and gene druggability information frompapers, andonline databases.
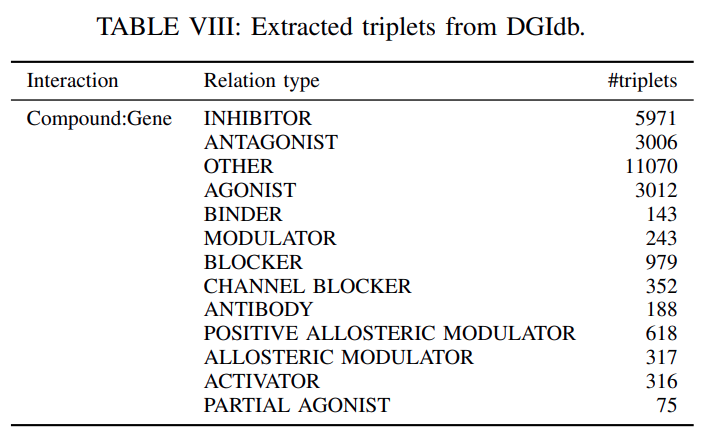
7)We extracted
26290 triplets from 13 relation typesas shown inTable VIIIand2551 gene entities and 6348 compound entities.
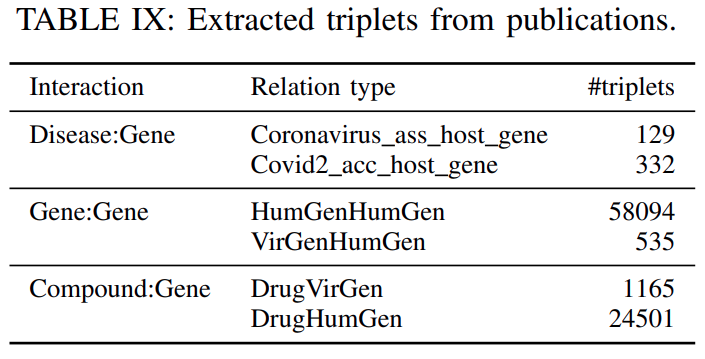
Data related to Covid-19:To further enrich our data with information related to Covid-19, we included interaction data from three recent publications(shown inTable IX).The work by Ge et. al.
developed a data-driven drug repurposing frameworkthatutilizes a biological network to discover the potential drug candidates against SARS-CoV-2. From that work, weextracted the biological network describing the interactions among host human proteins, virus proteins and chemical compounds.The proteins are indexed by the
UniProt IDand the chemical compounds by theirInChIKey.
In an effort to
discover antiviral drugsfor Covid-19, Gordon et. al.cloned,taggedandexpressed26 of the 29 viral proteins in human cells andidentified the physically associated human proteins.
They identified 67 druggable human proteins targeted by 69 existing FDA-approved drugs, drugs in clinical trials and/or preclinical compounds.
A framework for drug-repurposing is introduced by Zhou et. al., where the task is to repurpose drugs that are effective for
certain related coronavirus strainssuch asIBV,HCoV-229E,HCoV-NL63,SARS,MERSandMHV.From that work, we
extract relations among the aforementioned diseases and host proteins.
命名约定和规范化
From each dataset, we extracted a list of triplets in the form of
(head-entity, relation-type, tail-entity).
For representing entities, we usean entity type identifier followed by a unique ID of the specific entity, e.g., Gene::229475.
For representing relations, we usea naming convention that combines the name of the data source, the name of the relation, and the types of head and tail entitiesthat are involved e.g.,DGIDB::INHIBITOR::Gene:Compound.
Data sources use one of several ID spaces to represent genes, compounds, diseases and others.For example,
the same chemical compoundmay be represented in thedrugbank compound ID spacein DrugBank and in thechembl compound ID spacein the DGIdb.
To ensure that information from different sources in
integrating correctly, wemap biological entities to a common ID spaceusing the following rules:
Compound entitiesare mapped tothe drugbank compound ID spaceand if not possible tothe chembl compound ID space. If a compoundcan not be foundto either of the two we usethe native ID spaceand weinclude the name of the source as part of the entity’s name(e.g., Compound::brenda:1695336).
Gene entitiesare mapped tothe Entrez ID space.
Disease entitiesare mapped tothe MESH ID space.
The remaining biological entitiesappearonly in a single data sourceand hence we usethe data source’s ID.These rules are applied to
the biological entities per databasetomap the entities to the common ID space.
Finally, in order to avoid
relationsfor which we do not have enough data to train good embeddings, weexclude relations types that have less than 50 edges.
The DRKG – Putting everything together
The extracted set of
normalized tripletsfromthe previous datasetsconstitutethe Drug Repurposing Knowledge Graph (DRKG)that we created.It contains
97,055 entities belonging to 13 entity-types.The type-wise distribution of the entitiesis shown inTable X.

DRKG contains
a total of 5,869,294 triplets belonging to 107 relation-types.
Table XIshowsthe number of triplets between different entity-type pairs for DRKG and various data sources.The per-relation-type statistics have already been discussed in
Section III-A.
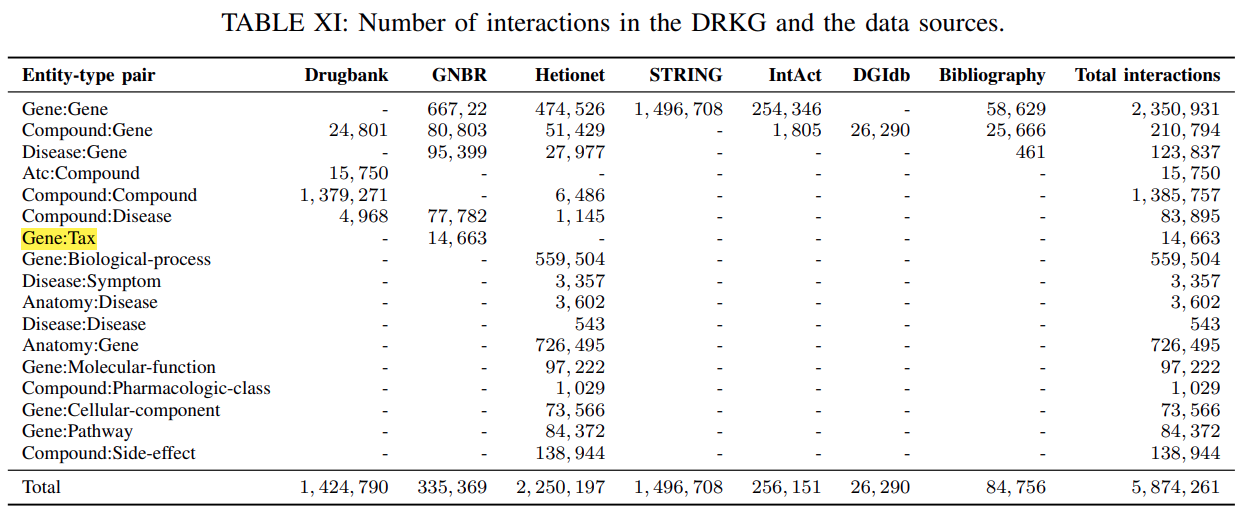
Figure 1depicts the possible interactions between the entity-type pairs in DRKG.
在 DRKG 上使用知识图谱嵌入进行药物再利用
Drug repurposing (DR)refers tousing existing drugs for new therapeutic indications.In this section, we
formalize the DR objective as a link prediction task over the DRKGthat can be solved byKGE models.In
the appendixwe includeseveral data analysis techniquestoverify that the constructed DRKG and the learned KG embeddings are of high quality.
Formulating drug repurposing as knowledge graph completion
DR refers to using existing drugs for new therapeutic indications.In the context of knowledge graphs,
DRcan be formulated as topredict new links between drug entities and disease entities of link type treat, orbetween drug entities and gene entities of link type inhibit or bind where the genes are related to the disease of interest(e.g.,involved in related pathways).In Section IV, we
validate DR on the DRKG for the Covid-19 disease, where we usethe direct link formulation of DR.By using our comprehensive DRKG, researchers can
address drug-repurposing for a variety of diseasessuch asHIVandSARS, as well as, the novelCovid-19.
Using knowledge graph embeddings for link prediction
Here, we
analyze DRKGby learning a TransE KGE model that utilizes the distance; see alsoSection II-B.By
optimizing equation (1)we obtainvector embeddings of dimensionforall biological entities and relationsparticipating in the DRKG.Consider the triplet of the DRKG and the associated score as
where is
a parameter of the TransE modelthat is set to12.0.For each triplet , the closer is to , the more confident the model is that the head and tail entities are connected under relation .
Drug repurposing using different relation types
In this section, we
evaluate our DRKG in the drug repurposing task for the Covid-19using the trained TransE KGE model inSection B.
Here we
use corona-virus diseases, includingSARS,MERSandSARS-COV2, astarget diseases representing Covid-19.We consider
two formulationsfor the DR task.
The first one predicts direct links between the disease entities and the drug entititesin the DRKG, whilethe second one predicts links among gene entities that are inhibited by drug entitieswherethe genes are associated with the target disease.We
select FDA-approved drugs in Drugbank as candidates, whilewe exclude drugs with molecule weight less than 250 daltons,as many of certain drugs are actually supplementsand we exclude them forsimplicity.This amounts to
8104 candidate drugs.We
collect 32 clinical trial drugs for Covid-19 to validate our predictionsandthe drug namescan be found inTable XII.

For
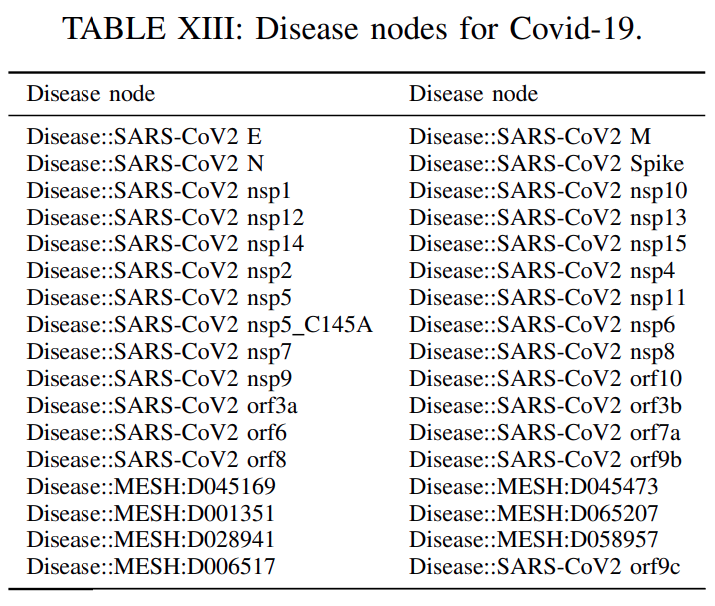
predicting links among disease and drugs with the relation treatment, we identifythe disease entitiesin our DRKG that are related withthe Covid-19 target disease.These disease nodes
constituteour expanded target set, since in certain cases asCovid-19, we may havemultiple disease nodes representing this novel diseaseor being related to it such asSARSandMERSdiseases.
The disease nodesforCovid-19are inTable XIII.
For this experiment, we select
‘GNBR::T::Compound:Disease’and‘Hetionet::CtD::Compound:Disease’as the target relations sincethese represent that a certain drug is used for treating a disease.Next, we
recover the pretrained embeddingsthat are obtainedusing the complete DRKGandfind the 100 drugs with the highest score using Equation (5).Finally,
to assess whether our prediction is in par with the drugs used for treatment, wecheck the overlap among these 100 predicted drugs and the drugs used in clinical trials.
Table XIVliststhe clinical trial drugs included in the top-100 predicted drugsalong withtheir corresponding score and ranking.
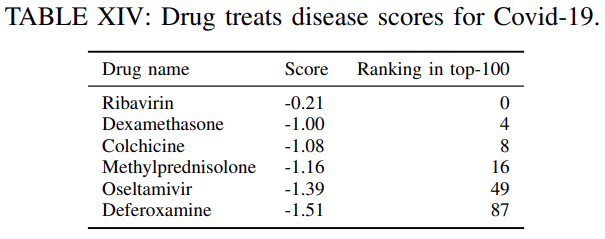
Evidently, using
the proposed DRKGandplain vanilla KGE model, several of the commonly used drugs in clinical trials are identified.
Finally on
Table XVwe reportthe top-10 highest ranked drugs for this experiment irrespective of whether these are used for clinical trials or not.
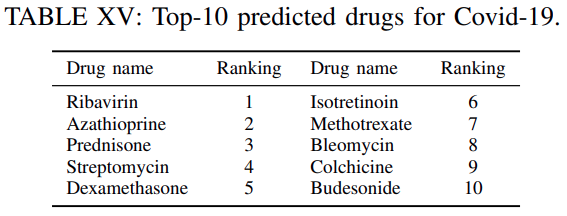
For
predicting links among gene and drugs with the inhibit relation, we identify the biological gene entities in our DRKG that are related withthe Covid-19 disease.
These gene nodesare involved inthe related pathways of Covid-19.We obtain
442 Covid-19 related genesfrom the relations extracted from [18], [9].
In this experiment, we select
the inhibit related relationwhich appears in three datasources as’GNBR::N::Compound:Gene’,’DRUGBANK::target::Compound:Gene’and’DGIDB::INHIBITOR::Gene:Compound’.We compare
the results obtained by the DRKGwiththe corresponding resultsif wetrained a KGE on a subset of the DRKG that uses only relationsfrom the three databases GNBR, DRUGBANK and DGIDB individually.
For the results, we
recover the pretrained embeddingsthat are obtainedusing the KGE modelandfind the 100 drugs with the highest score using Equation (5)and rank them per target gene.This way we obtain 442 ranked lists of drugs.
Finally,
to assess whether our prediction is in par with the drugs used for treatment, we check theoverlapamongthese 100 predicted drugsandthe drugs used in clinical trials per gene.This procedure is
repeated three times per relationand we comparethe results of using the DRKG against the constituent databasesthat include an inhibit relation.
Tables XVI-XVIIIlistthe clinical drugs included in the top-100 predicted drugsacross all the genes withtheir corresponding number of hitsfor DRKG and the constituent databases.
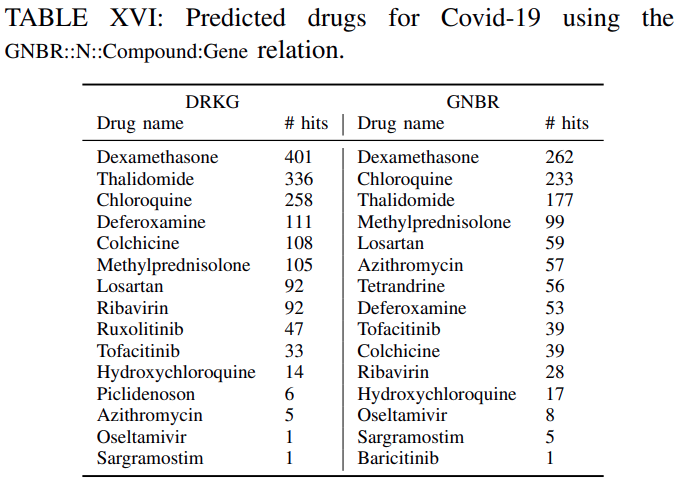

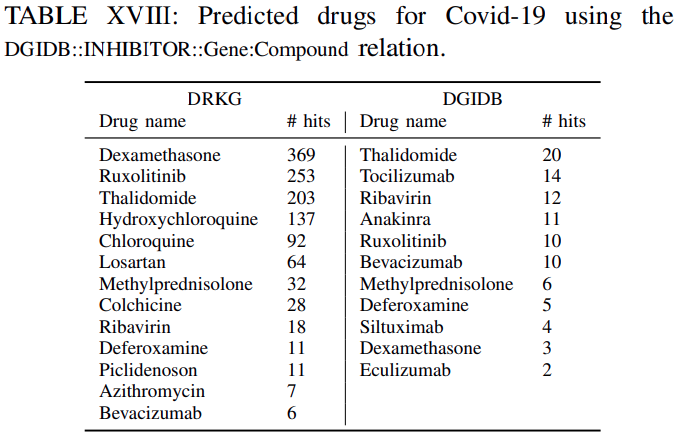
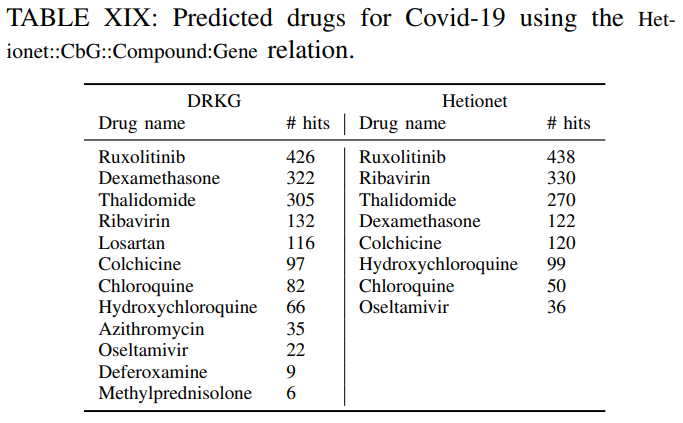
The number of hits shows in how many gene ranked lists the suggested drug appeared in the top-100 ranked drugs.For example ifa drug appears in the top-100 ranked drugs for all genes the number of hits is 442. This isthe maximum number of possible hits.It can be observed, that
several of the commonly used drugs in clinical trials appear high on the predicted list.Furthermore,
the number of hits using the DRKG is significant highercomparing tothe constituent databases, which corroborates themerits to constructing a comprehensive DRKG.
结论
This paper constructed a
DRKGfrom a collection of data sourcesthat can be utilized for general drug repurposing tasks.To further facilitate efforts of researchers in
repurposing drugs for Covid-19we also include in DRKGproteins and genes related to Covid-19, as extracted fromrelevant papers.We train
KGE models on the DRKGand obtainembeddings for entities and relation types.We also validate that the DRKG structure and the learned embeddings are of high quality.
Finally, we evaluate the DRKG in the drug repurposing task for Covid-19.
It is observed that
several of the widely used drugs in clinical trials are identified by our method.
Our future research effortswill focus onincluding more biological entities in the DRKG,enhancing the entities with attributessuch aschemical sequence for compoundsanddeveloping deep graph learning modelsthat are dedicated fordrug repurposing.
附录
This section describes data analysis techniques we performed on the DRKG. The goal is to verify that the constructed DRKG and the learned KG embeddings are of high quality. The code is implemented in DGL-KE that is an open-source package to efficiently compute knowledge graph embeddings. It contains several KGE models, including TransE, DistMult and RotatE, and introduces various optimizations that accelerate training on knowledge graphs using multi-processing, multi-GPU, and distributed parallelism. This way DGL-KE facilitates efficient training on knowledge graphs with millions of nodes and billions of edges.
Graph structure analysis
Here the quality of the constructed DRKG is assessed. Different data sources may describe the same triplets and hence the constructed DRKG may have some redundant edges. In this section we verify that triplets from different sources do not have significant overlapping information.
First, we assess what is the percentage of common triplets among each pair of edge types in the DRKG. This is important since the same relation-type among the same nodes might be described in two different datasets, and the DRKG may over-represent this relation. If the percentage of common triplets is relatively small then combining these is well justified. Towards this end, we compute the Jaccard similarity coefficient among a pair of relation types , that is defined as follows
where denotes the cardinality of the set . The Jaccard score is a measure of similarity among the relation types, if is close 1 then and are connecting the node pairs. Table XX reports the 10 most similar edge-pairs based on their Jaccard coefficient. As expected some relation types connect the same nodes since the information from different data sources is related. Nevertheless, the relative small values of the Jaccard coefficient for the most similar edge-type pairs indicates that there is value in including the edge types from all the sources. For example, enzyme catalysis is typically done through binding interactions with protein substrates directly or indirectly.
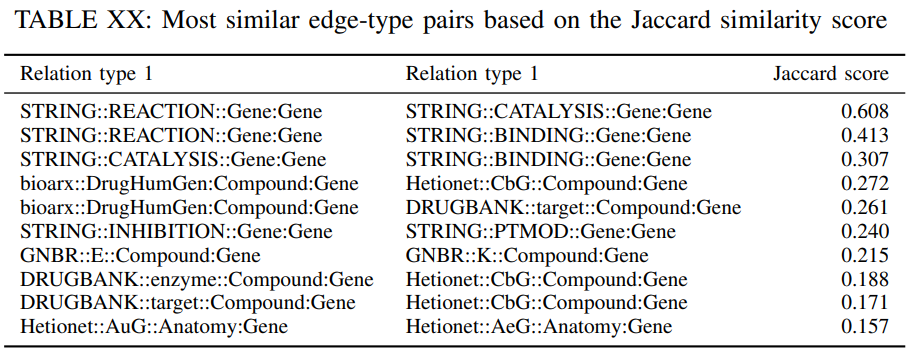
In our DRKG, the relations “STRING::REACTION::Gene:Gene”, “STRING::CATALYSIS::Gene:Gene” and “STRING::BINDING::Gene:Gene” have high Jaccard similarity values (0.608, 0.413, 0.307), that supports our understanding of these activities. The three relations “bioarx::DrugHumGen:Compound:Gene”, “Hetionet::CbG::Compound:Gene” and “DRUGBANK::target::Compound:Gene” from different databases correspond to a same relation that compounds bind to their target genes, and in our DRKG, these relations have high pairwise Jaccard similarities. Compounds could affect gene expression after they are metabolized into active substances and moved to certain organs where the genes are usually highly expressed, or compounds and genes compete for same enzymes. In our DRKG, the high Jaccard similarity between “GNBR::E::Compound:Gene” and “GNBR::K::Compound:Gene” corresponds to such a biological process. The relations “Hetionet::AuG::Anatomy:Gene” and “Hetionet::AeG::Anatomy:Gene” both represent that the gene is over-expressed in the Anatomy, except for AuG in post-juvenile adult human samples. In our DRKG, they have high similarity under Jaccard.
The Jaccard similarity may not capture the case that an edge type is contained in another one but the two edge sets have significantly different sizes. Nevertheless, this could happen if two databases describe the same relation, but one has significantly less edges than the other. Next, we examine whether all the edges of a certain type are described by another edge type as well that is . To accomplish this, we compute the overlap coeffcient that is defined as
The overlap coefficient is close to 1 if all the edges in one edge set are also present in the other set. Table XXI reports the 10 most similar edge-type pairs based on their overlap coefficient.
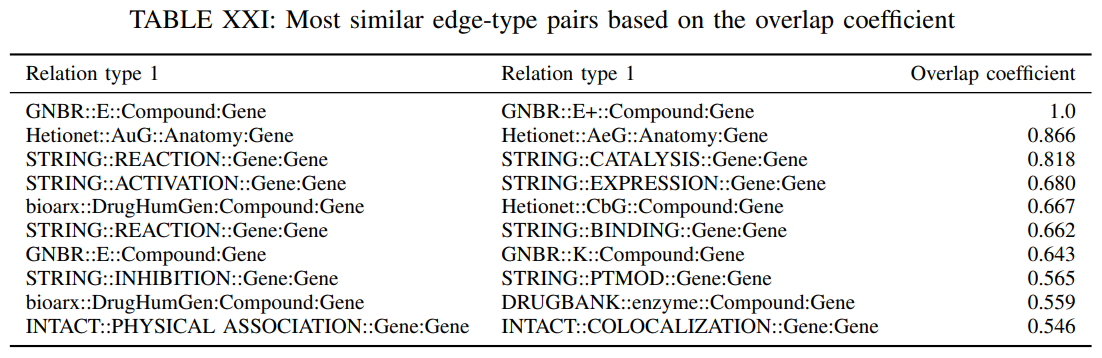
We observe that for certain edge-type pairs there exists significant overlap. Nevertheless, for the pair GNBR::E and GNBR::E+ the total overlap is expected since the first relation signifies that a drug affects the expression of a gene whereas the second indicates that a drug increases the expression of a gene and it is contained in the first one. The high overlap coefficient between “Hetionet::AuG::Anatomy:Gene” and “Hetionet::AeG::Anatomy:Gene” is also expected, because AuG is a special case of AeG. Similarly, “STRING::CATALYSIS::Gene:Gene” and “STRING::BINDING::Gene:Gene” can be considered as special cases of “STRING::REACTION::Gene:Gene”.
Knowledge graph embedding analysis
We can use the embeddings of the knowledge graph to analyze how relations and entities are clustered and investigate whether certain similarities in the embedding space are consistent with their biological meanings, e.g., “GNBR::E:Compound:Gene” and “GNBR::E+:Compound:Gene” can be similar as they both represent the expression relationship between a Compound and a Gene, but “GNBR::E+” represents the positive expression. Further, we can generate the confidence of existing edges by calculating the prediction scores using the embeddings. From the perspective of the knowledge graph embedding algorithm, e.g., TransE, if the score between two entities and under relation is far away from , then the model cannot correctly score an edge on which it trained on. This means that the edge does not fit the underlying model and is often an indication that the corresponding edge may be incorrect.
Here, we analyze DRKG by learning a TransE KGE model that utilizes the distance (Section II-B). For the analysis in this section, we split the edge triplets in training, validation and test sets as follows 90%, 5%, and 5% and train the KGE model. Finally, we obtain the entity and relation embeddings for the DRKG. Next, we apply the following methodologies to validate the quality of the learned embeddings.
1) Entity embedding similarity: We use t-SNE to map entity embeddings to a 2D space. Figure 2 illustrates how the entity embeddings are placed in the 2D space. Different colors denote different entity types. We observe that entities from the same type are grouped together as we expected.
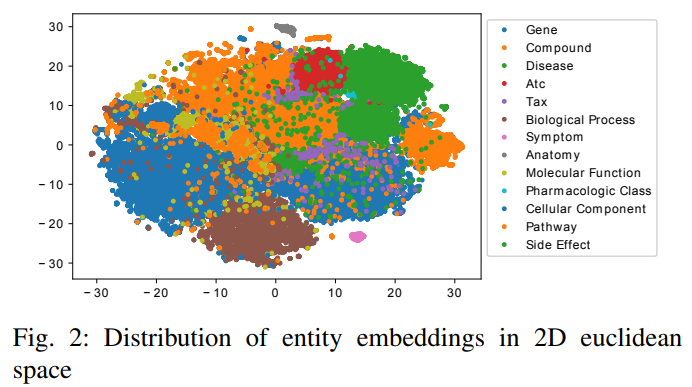
Table XXII shows the average pair-wise cosine similarity of certain entity type within the same category and cross different categories. It can be seen that entities are more similar to each other within the same category that entities from different categories.
Figure 3 shows the detailed distribution of pairwise cosine similarity between different entities based on their embeddings. In the figure, the counts are normalized that the area under the histogram is sum to 1. It can be seen that most of the entities have low cosine similarity according to their embeddings. They are distinguishable in the current embedding space.
2) Relation type embedding similarity: We use t-SNE to map relation embeddings to a 2D space and plot it in Figure 4. It can be seen that relations are widely spread across the 2D space and relations from the same dataset do not cluster which is expected as most of relations have different meanings even from the same data source.
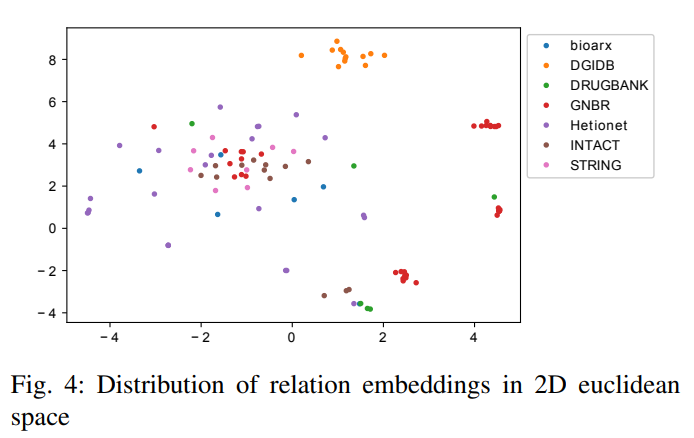
Only a small part of relations from GNBR dataset are clustered together. There are two clustered red dots (relations from GNBR) in the Figure. Table XXIII shows all relation-type pairs with the Cosine similarity larger than 0.9. It can be seen that “GNBR::E::Compound:Gene”, “GNBR::K::Compound:Gene”, “GNBR::E+::Compound:Gene”, “GNBR::N::Compound:Gene” and “GNBR::E-::Compound:Gene” are highly similar to each other.
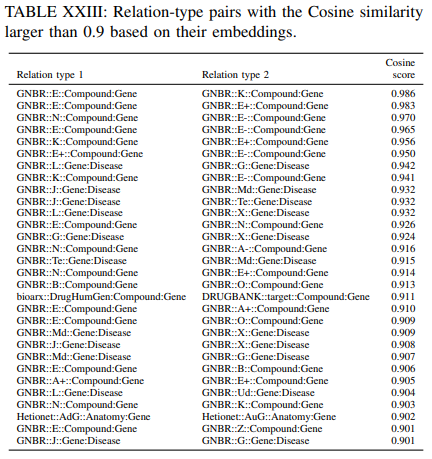
Some of these pairs also appear in the most similar edge-type pairs analysis on the Jaccard similarity and overlap similarity. Drugs can affect (E) gene expression (either increase (E+) or decrease (E-)) through their pharmacokinetics (K). Drugs can also inhibit genes to affect gene expressions and thus the high cosine similarity between “GNBR::N:: Compound:Gene” and the above relations are justified. “GNBR::L::Gene:Disease”, “GNBR::G::Gene:Disease”, “GNBR::J::Gene:Disease”, “GNBR::Md::Gene:Disease”, “GNBR::Te::Gene:Disease” and “GNBR::X::Gene:Disease” are highly similar to each other. The similarity between these relations could be due to that the genes are involved in the pathogenesis of the disease and therefore they can be treated as the targets or they have therapeutic effect for the disease. From the table, we can see that “bioarx::DrugHumGen:Compound:Gene” and “DRUGBANK::target::Compound:Gene” are also similar to each other, while they have similar meaning of treatment.
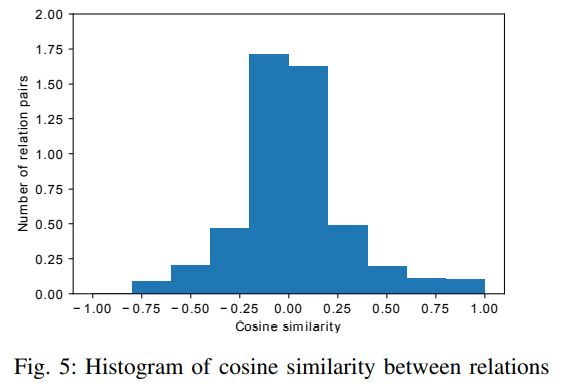
Figure 5 shows the detailed distribution of pair-wise cosine similarity among different relation types based on their embeddings. The counts are normalized that the area under the histogram is sum to 1. It can be seen that most of the relation embeddings have small cosine similarity. Only 0.53% of relation pairs have similarity larger than 0.9 with the maximun of 0.986 (between GNBR::E::Compound:Gene and GNBR::K::Compound:Gene).
3) Edge prediction analysis: Towards validating each triplet of the DRKG we evaluate how well it fits the scoring function of KGE model. In order to avoid the possible bias of over-fitting the triplets in the training set, we split the whole DRKG into 10 equal folds and train 10 KGE models by picking each fold as the test set and the rest other nine folds are the training set. Following this, the score for each triplet is calculated while this triplet was in the test set. Consider the triplet of the DRKG and the associated score as
where is a constant we used in training of the TransE model that is set to 12.0. For each triplet , the closer its score is to , the more confident it was that the head and tail entities are connected under relation . The distribution of the edge scores is depicted in Figure 6.
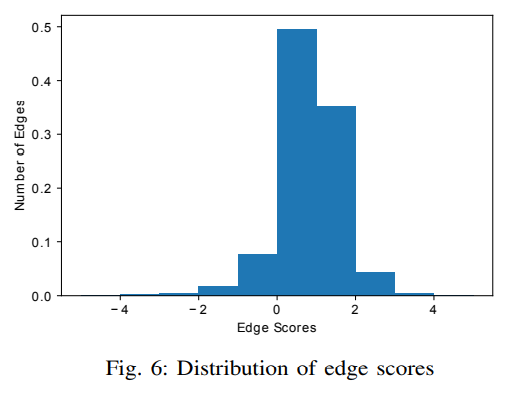
The counts are normalized that the area under the histogram is sum to 1. From the figure, we can see that around 57.28% of edges are scored as |score| < 1 and 94.24% of edges are scored as |score| < 2. The average of |score| is 0.987. We also randomly shuffle in for all triplets to construct the negative triplets and calculate the using the same formula. The average is 2.545.
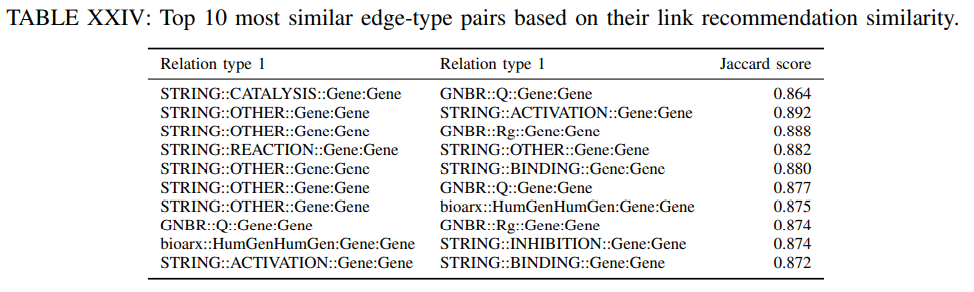
4) Link type recommendation similarity: Finally, we also evaluate how similar are the predicted links as given by the KGE model among different relation types. This task examines the similarity across relation types for the link prediction task. For a set of nodes, we measure the overlap of predicted neighbors under different relation types as given by the TransE model.
For seed node we find the top 10 neighbors under relation with the highest link prediction score. Next, we repeat the same prediction for relation and calculate the Jaccard similarity coefficient among the predicted sets of top 10 neighbors for and . We repeat this process for 100 random selected seed nodes and report the average similarity score for all edge-type pairs. The most similar edge types in this context are the ones relating Genes. This may be attributed to the fact that the genes are the most represented entitites in the DRKG.
结语
第四十一篇博文写完,开心!!!!
今天,也是充满希望的一天。














